The Seung Detector AKA museum.eyewire.org
Welcome to the future! We have flown past Pluto. Google Loon balloons will soon supply all of Sri Lanka with internet. And now gamers like you are discovering new types of neurons.

We’re pleased to announce a public alpha prototype release of a new scientific tool and visualization platform made possible thanks to EyeWire: The Seung Detector, or as it’s known at Princeton, The Cell Museum. Explore it at museum.eyewire.org and check out the description below so you’ll know what you’re seeing. It’s used for – you guessed it – classifying and detecting cell types.
Countdown to What
On July 27, 2015, the EyeWire community completed the Countdown to Neuropia, a systematic reconstruction of all 245 neurons within a large region of the entire EyeWire dataset. Thus begins our quest to create the world’s most comprehensive catalog of neuron types in the retina. Mapping neurons is only part of the scientific process. Next we need to analyze the models to understand which cells connect and differentiate types and classes.
Many of the neurons we mapped during the countdown are novel — they’ve never been seen before! To be certain that they are new, we need to find more examples of the unique neurons, hence Bonus Mystery Cells in EyeWire.
How to use the Seung Detector
What have we have discovered so far? Without a little analysis, we’ll never know! That’s why we’re building the Seung Detector, a state of the art tool that showcases different neuron types alongside data, allowing classification.
We encourage you to explore an alpha version of this software, created by Igancio Tartavull* at Princeton, at museum.eyewire.org. It features gallery views where, for the first time, you can see multiple cells from EyeWire in the same window.
Select from the navigation bar on the left until you get to a window that looks like this – there are so many neurons to explore!

Protip: click the cell ID number beneath the stratification graph on right to hide that cell from view. Fewer cells = faster loading.
Minimize the menus/maximize the neuron view by selecting the double sided arrow on the cell view window. Scroll to zoom.
There are quite a few bizarrely named items to choose from. Before you dive in, you might want to know what those names mean.
Neuron Naming Abbreviations
What’s this mess about “gc bn1d” or “gc bw3rw,” you might wonder? These names shorten names that would otherwise be totally ridiculous, such as “bistratified wide regular ganglion cell of depth 3.” Yeah.
Your key to scientific sanity:
gc/ac = ganglion cell/amacrine cell
Many of the neurons we mapped during the countdown are “ganglion cell” type, which means they are found in the ganglion cell layer of the retina. We also mapped amacrine cells, which are greater in number and will be analyzed after the ganglions.
m/b/t = monostratified, bistratified or tristratified
a = asymmetric
Not all cells are branched equally! Dendrites of some cells concentrate on one side, making them asymmetric.
1-10 = depth in retina
r/d = regular or diffused
Some neurons have concise layers while others are more gnarly!
n/w = narrow or wide
Does the arbor (branched region) have a small diameter or a large one? Breadth in lateral size.
s/t = short or tall
Does the arbor occupy a shallow region of the dataset? Breadth in depth.
o/f = on or off
EyeWire cells’ branches are found in a region of the retina called the inner plexiform layer. This layer is further subdivided into two halves: the on and off layer. Cells in the on layer are activated when photoreceptors detect light. Can you infer how cells in the off layer are activated?
Bistratified cells have one layer of dendrites in O Layer and another layer of dendrites in F Layer. This descriptor on/off is not relevant for monostratified cells.

Note that some of the letters in naming are modifiers that help distinguish cell types with the same depth number. For example, tall/short and narrow/wide are only relatively true within the types that share the same number. mXw is wider than mXn but it can be narrower than mYn. If two cells have similar stratification, the one that stratifies closest to the ganglion cell layer (GCL) is i (inner) while the one farther from the GCL is o (outer).
To The Future
Over the coming months, we will add numerous new features, more data, and even a way for the EyeWire community to help classify cells.
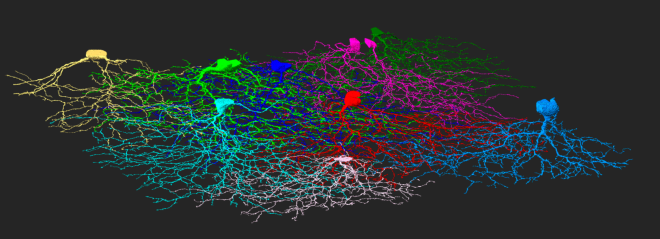
For now we hope you enjoy exploring The Seung Detector. We’ll leave you with a circuit of neurons you discovered during the Countdown to Neuropia. Look closely and you might even see a few synapses 🙂
Thanks for playing and we’ll see you online at EyeWire.org.
For Science!

* Additional contributors to museum.eyewire: Jinseop Kim, Sebastian Seung, Chris Jordan, Will Silversmith, Mio Akasako